Software
Sequence software
Benedikt Poser and members of his group are well known in the field for his sequence developments. The most-popular sequence implementations are:
- Mutli-echo EPI readouts for artifact reductions in fMRI.
- 3D-EPI readouts for high temporal and/or spatial resolution fMRI.
- Simultaneous-Multi-Slice (SMS) imaging for coverage increases in fMRI.
- Parallel transmit tools at high magnetic fields.
- Advanced functional readouts for VASO and ASL imaging.
These tools have partly been included into SIEMENS work-in-progress (WIP) packages and can be requested directly from SIEMENS.
Other sequences have been widely shared via SIEMENS C2Ps and are available upon request.

Users of the sequence technologies developed by the MR-Methods group.
High-resolution processing scripts
These are a bunch of wrappers and scripts for accurate analysis of ultra-high-resolution fMRI. Written by Sriranga Kashyap and developed for unconventional datatypes and rely on the ANTS. It’s has been used for its routines of ultra-high resolution alignment across runs/days and the integration of motion correction, distortion correction, and co-registration into one resampling step etc. The code will be made available via Gitlab.
MBIC-MR-Methods
Group GitHub: https://github.com/MBIC-MR-Methods.
LAYNII
LAYNII is a collection of C++ programs designed for layer-fMRI analysis and VASO analysis. LAYNII is mostly developed by Renzo Huber and used code snippets from the AFNI-group for the nifti I/O. The name refers to analysis of LAYer-fMRI with NIfti data. LAYNII that operates exclusively in voxel-space, not in vertex space. LAYNII is designed to generate topologically consistent estimated of layers and columns with partial brain coverage data.
It can be downloaded and installed from Github.
Step-by-step instructions and example cases are provided as layerfmri.com.
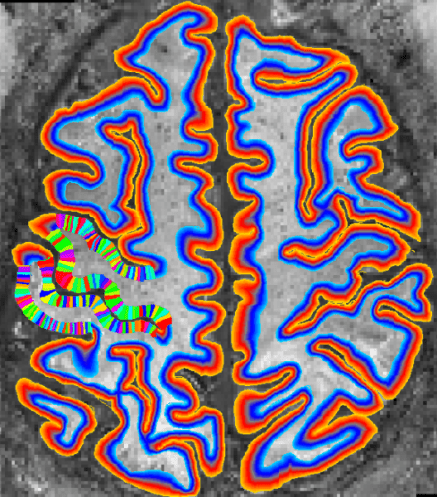
The most-often used programs of LAYNII are tools to estimate layers and columns for unfolding of GM patches.
Recon Tools
The MR-Methods group uses and developed advanced reconstruction tools. Specifically Gilad Liberman developed Minimal Linear Networks (MLN) for the reconstruction of spiral readouts.

Example result of MLN reconstruction with reduced artifact levels.
The corresponding software is available on Github.
